We develop and use microfluidic devices to explore questions in biology. Often this involves making picoliter to nanoliter aqueous droplets in an inert, immiscible, carrier fluid. Surfactant keeps each droplet separate from every other droplet, so we can treat each as a reaction vessel containing an independent experiment. This gives us massive throughput --- if we make one-million drops, that is in principle one million separate experiments. This approach excels at high-throughput screening; for example, if we encapsulate a single cell in each drop along with PCR or other DNA/RNA amplification machinery targeting specific mutations, we can quickly screen millions of cells for the presence of the mutation of interest. This already been used to quickly characterize and find rare mutations in tumor cells, enabling personalized treatments. We are also not limited to directly screening the genome; we also can screen for desirable phenotypes. Currently, we use phenotypic screening to both search for cells that produce specific antibodies, and as a key component in an automated system to directly evolve enzymes.
In another powerful biological application, droplet microfluidics provides ability to study cell-cell or cell-microbe interactions. In this scenario, we encapsulate multiple cells or microbes in each droplet and observe how the cells interact. For example, we encapsulate bacterial and mammalian cells in droplets to understand the difference in how pathogenic and non-pathogenic bacteria interact with mammalian cells. In the past, we have also used this approach to study the interaction between single viruses and mammalian cells. In studying interactions, the power is again in the throughput. For a given timeframe, we can perform over a thousand times more cell interaction experiments using a droplet-based approach than is possible using traditional well-plate methods.
As a final example, we use droplet-based microfluidics to perform single-cell transcript sequencing and we develop new single-cell DNA and RNA sequencing platforms. Since droplet-based highly parallelized single-cell transcript sequencing was introduced by this lab in 2015, is has become the standard approach, and has enabled the recent single-cell data mining "goldrush." The ability to distinguish and quantify transcripts from thousands of individual cells within large cell populations and tissues has provided invaluable insight in a number of different biological disciplines, such as developmental biology, tissue engineering, cancer biology, and even microbiology.
These are just three examples of some of the work we do at the intersection of microfluidics and biology. We are always interested in new ideas and trying new things. To learn more about what is currently going on in the lab, please see the research projects below!
Here are some current projects in our group:
|
Microfluidic droplets to identify and isolate individual desired immune cells: Microfluidic water-in-oil droplets can be formed, observed, and manipulated at ~kHz rates. Mammalian cells retain activity within these droplets, and, due to the small droplet volume (~50 picoliter), products secreted from single cells can reach detectable levels after <1hr incubation. We exploit these features to develop in-droplet assays to identify and isolate individual cells of interest. John Heyman |
|
Modelling the Human Gut Microbiome in Droplets: The human gut microbiome is extremely diverse. Different microbiota can interact with pharmaceutical drugs in different ways. A model of the gut microbiome inside microfluidic droplets would allow us to test drugs at very high throughput either by screening many drugs through a single microbiome or a single drug through many microbiota. Currently we are attempting to do this by growing intestinal epithelial cells on the inside of a alginate gel capsule and then filling the inside with bacteria. Ben Thorne |
|
 |
Understanding bacterial persistence using single-cell transcriptomics: Antibiotic persistence is a global public health problem wherein a tiny fraction of any bacterial population - in the context of bacterial infections that's pathogenic bacteria - manages to survive even the most lethal doses of antibiotic attack, lie low during the antibiotic treatment, and then start to multiply once the antibiotic is flushed out of their environment. This behavior, which is entirely due to a difference in the gene expression of the persister cells as compared to the non-persister cells, often causes relapse of bacterial infections in the clinical context, and predisposes the persistent cells to develop the more fearful antibiotic resistance. In my research, using single-cell RNA sequencing, imaging and other more classical microbiological assays, I am trying to understand and discover the different biological mechanisms that the persistent cells employ to become persisters. Debojyoti Panda |
|
Mechanical Lysis of Single Bacteria: We are working on a mechanical method to break the cellular membrane of individual bacteria, also known as lysis. Single-cell analysis is incredibly important as it allows us to study rare populations and understand the differences between single bacteria in a large population. Lysis is the crucial step, and current chemical methods to carry out single-cell lysis limit the downstream applications. In contrast, this method is mechanical; it uses a traveling surface acoustic wave (TSAW) within a microfluidic channel to lyse each single bacteria separately. Kayla Keepseagle |
|
|
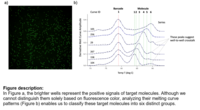
Programmable multiple temperature curves for barcoding massive and versatile bioassays: Biomarkers are measurable indicators of biological processes, conditions, or diseases in the human body. They can be classified into genetic, proteomic, and metabolomic biomarkers based on their origin and function. DNA and proteins are two primary types of biomarkers that play vital roles in healthcare and medical research. They enable early diagnosis of diseases and help physicians identify appropriate treatment courses. Additionally, they can be applied to assess the safety and efficacy of new drugs, accelerating drug discovery, and reducing costs. In this study, we utilize the melting temperature of DNA as the readout signal, representing the temperature at which half of the double-stranded DNA (dsDNA) becomes single-stranded (ssDNA). By adding a melting temperature barcode to target molecules, we can generate a unique melting curve pattern for each target molecule. These distinct melting curve patterns can be used as fingerprints for the specific identification of each targeted molecule. Moreover, by combining this approach with digital PCR, we can absolutely quantify the targeted molecules. Xing Zhao |
|
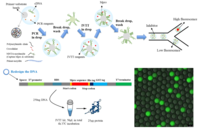
High-Throughput Screening of Drug-Resistant Viral Protease and Acid-Tolerant Glutenase using Droplet Microfluidics: Enzymes are functionalized proteins that act as catalysts, accelerating chemical reactions in biological systems. The enzyme evolution, which refers to the enzymes change in their sequence, brings the enzyme benefits such as tolerances to high temperature, acid, base and strong salinity, but sometimes the declines on targeting, binding and catalyzing efficiency. Our goal is to harness the potential of high-performing enzyme mutants while also elucidating the underlying mechanisms and strategies to improve less efficient enzymes. Here, we develop a high-throughput screening assay that combines droplet microfluidics with direct enzyme evolution. We produce a mutation library over 106 and screen out the enzyme mutants we are interested in. Specifically, we have applied our technique on the screening of drug resistant mutants of the SARS-CoV-2 (virus caused the COVID19) Main-protease (Junming Lao, Karla Durdic). Besides, we have promoted the evolution and identification of gastric acid-tolerant glutenases, offering a potential therapeutic avenue for individuals with gluten allergies based on this technology ( Zhang, Xinge (Diana), Junming Lao). |
|
Microfluidics fabrication of photothermal targeting dual responsive dextran-calcium alginate hydrogel for mRNA-based combination therapy: We develop water-in-oil emulsion droplet templates to encapsulate therapeutics including mRNA, antibody and anticancer drugs for targeted drug delivery with smart materials (porous silicon nanoparticles based nanocarriers co-loading hydrophobic and hydrophilic therapeutics with high loading capacity and photothermal targeting properties) on a PDMS microfluidics chip. The biocompatible functionalized calcium alginate hydrogel is fabricated through crosslinking with another droplet directly. The photothermal targeting dual responsive hybrid nanohydrogel could be developed through extrusion technique for mRNA-based therapeutics co-delivery and photothermal targeting double combination therapy. Mingtan Hai |
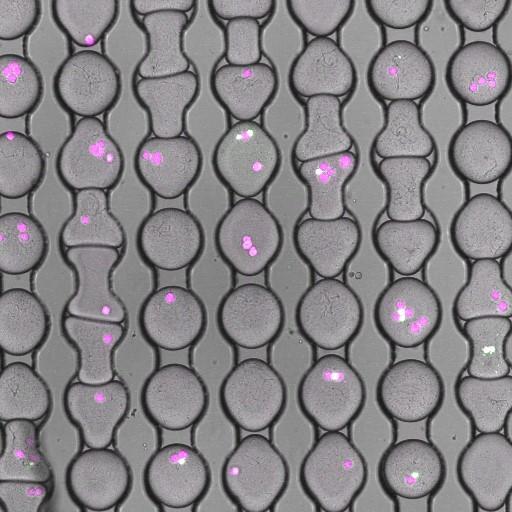
| Classifying metagenomic bacterial samples phenotypically based on traits that are essential for host colonization may help us in finding common genetic or transcriptomic signatures or mechanisms across different species that cause such traits, irrespective of whether we know their identity or not. In addition, the behavioral segregation beforehand may help us in discovering rare microbes by potentially cleaning up the sequence space during metagenomic analysis. Hence, we are developing bead-based ultrasensitive assays to classify known or unknown bacteria phenotypically based on the trait of bacterial adhesion to different human cells and extracellular matrix proteins. Furthermore, we are developing bead-based microfluidic platforms to study the adhesion and evolutionary dynamics of multi-species biofilms on complex substrates with tunable surface properties. Debojyoti Panda | |
|
High-sensitivity whole-genome sequence for discovery of new virus in sewage water: Environment is rich in microbial resources, and there may be as many as 1012 microbial species worldwide. Most of these microbes are not culturable or are not well understood. Metagenomic sequencing is a powerful culture-independent approach to discover novel, uncultured microbial and viral genetic diversity, which sequence the whole genome of all microbes in the environment. However, current metagenomic and bioinformatic methods typically miss important viral populations, mainly owing to biases and limitations in sampling or assembly steps. Single-virus genomics (SVG) is a new approach that is complementary to metagenomics. SVG can uncover uncultured, abundant and cosmopolitan viral populations, which may be lost by using the metagenomic sequencing method. The key advantage of microfluidic droplets is single cell encapsulation, manipulation, enrichment, detection and sorting. It is direction for genome recovery is to isolate single viruses experimentally and prepare individual sequencing libraries by microfluidic droplets. Single cell sequencing technology provides support for obtaining the genomes of rare species and discovering new metabolic pathways. Yi Xiao |
Here are some previous projects in our group:
|
A high-throughput approach to detect pathogenic bacteria: Identifying pathogenic bacteria (bacteria that cause diseases in humans) in natural samples has important consequences for human health. For example, 80% of foodborne illness comes from "unknown origin" --- there are either too few pathogenic bacteria in the sample to detect, or there does not exist a specific test for the pathogen at fault. To address these concerns, we focus on high-throughput methods that require no prior knowledge of the bacteria to assess its pathogenicity. Our current approach relies on droplet microfluidics: we co-encapsulate human tissue with the bacteria of interest, forming a functional assay capable of screening 106 bacteria per day. Perry Ellis |
|
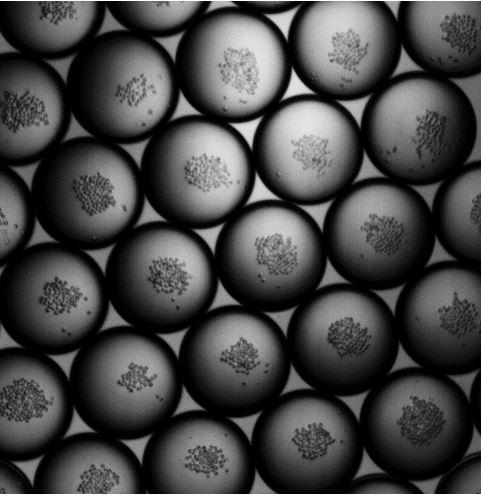
Better organoids through homogeneity: Organoids are 3-dimensional masses of cells that better reproduce in vivo function than tradition 2-dimensional cell culture. They currently show promise as sources of therapeutic cells, as better testing platforms for new drugs, and as platforms to better understand organ development in general. However, one problem with current protocols to produce organoids is the large variability in the mature organoids. This variability makes it difficult to do quantitative studies with organoids and consequently, realize their full potential. We are attempting to solve this problem using droplet microfluidics. The basic idea is to specify the initial conditions for each individual organoid such that they all identical at the start of their development. The hope is that a homogeneous starting condition leads to a more homogeneous mature organoid population. To get a homogeneous starting population, we randomly encapsulate starting cells into aqueous droplets and then sort for the droplets with the desired number and composition of starting cells. Currently, we are working on both the microfluids and the biology for this initial step. Perry Ellis |
|
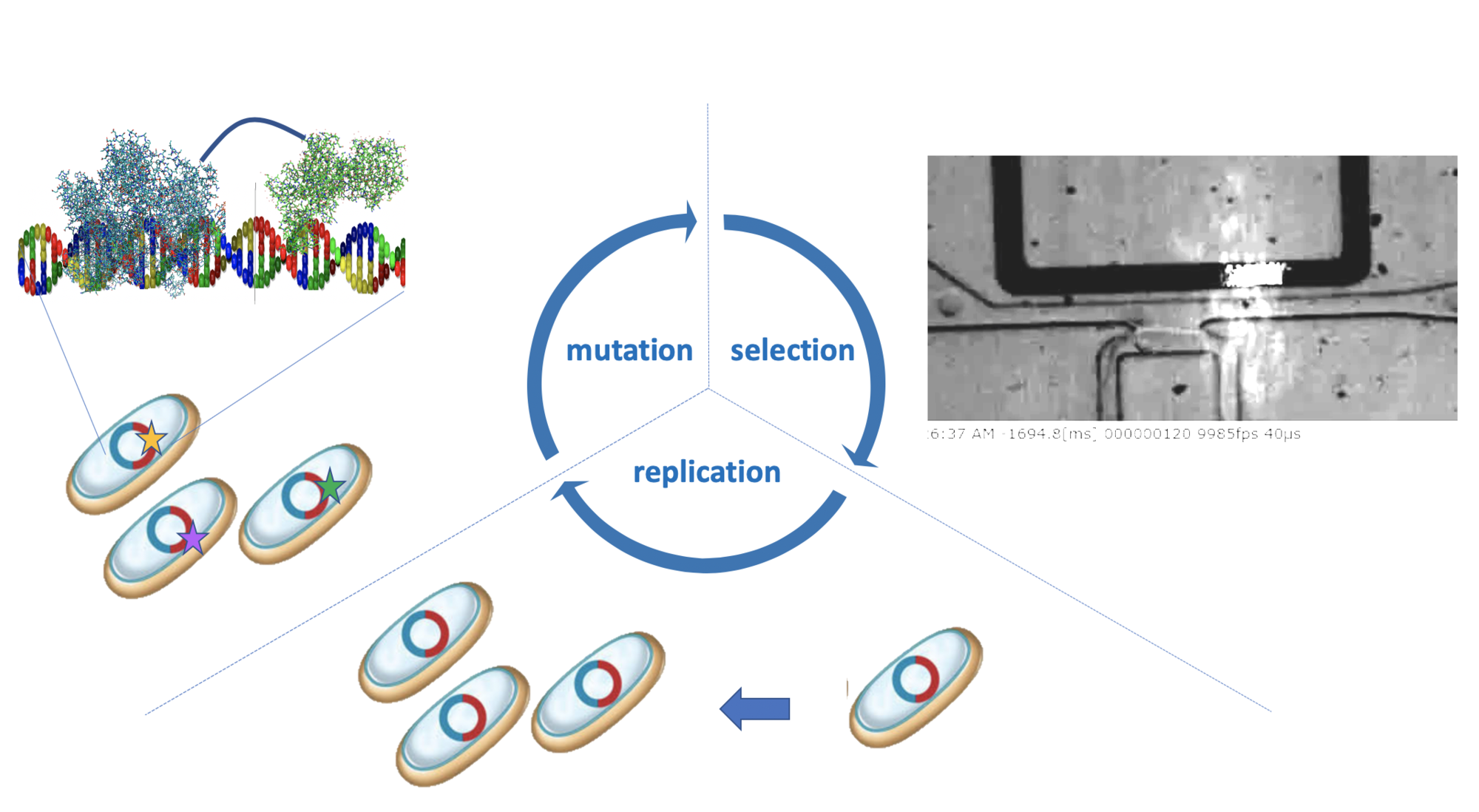
Machine-learning-guided directed protein evolution: Engineering protein to achieve certain functions is a necessary step in the development of many scientific and medical applications. To enhance desirable protein features, directed evolution is a reliable method, where beneficial mutations are identified and accumulated through an iterative protocol of mutation and selection. The selection process entails screening of hundreds to thousands of variants in each generation, which is a significant experimental burden. To accelerate this process, we combine machine learning methods and microfluidic technology to select highly potential variants and perform efficient screening for the target protein function. Machine learning enables us to model protein sequences and explore mutation effects. With these models, the performance of variants can be predicted. By selecting variants with high probability of having the desirable function, we greatly reduce the number of variants needed to be screened. In addition, we develop a droplet microfluidics system that allows fast and automated screening of variant activities, further reducing the time and labor of directed protein evolution. Liyin Chen |
|
High throughput screening of biological phenotypes is essential to better understand the genotype to phenotype linkage in living systems. We use single cells microfluidic assays to characterize the enzymatic activity of millions of single cells and create large scale activity to genotype mappings for machine learning approaches. Additionally, we are developing single cell continuous directed evolution approaches to leverage new genetics and high throughput screening to improve antibodies and enzymes. Raoul Rosenthal |
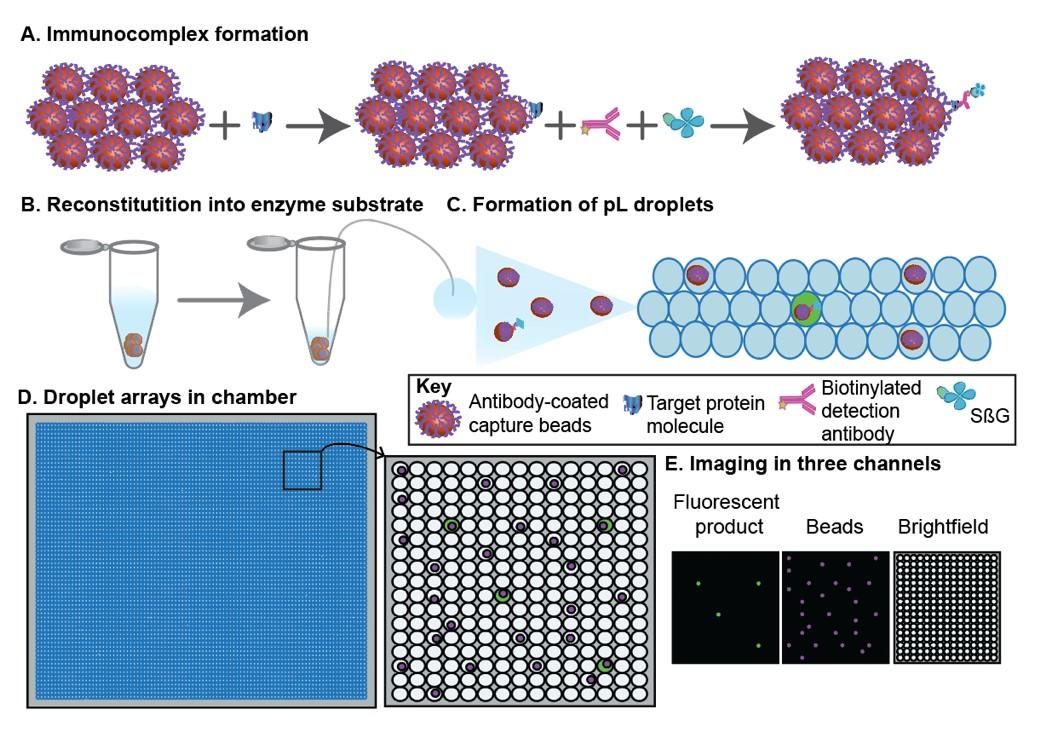 |
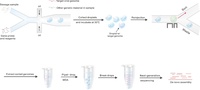
Ultra-sensitive protein detection using droplet digital ELISA: We developed a novel approach for ultra-sensitive, single-molecule detection of proteins using digital ELISA and droplet microfluidics (Figure1). It is a bead-based digital immunoassay in which beads are isolated in pL-sized droplets and then loaded into a chamber, forming droplet arrays, for analysis. More specifically, antibody coated paramagnetic beads are added to a sample containing the target molecule. The target molecule is then labeled with a biotinylated detection antibody and streptavidin-β-galactosidase (SβG), forming an enzyme-labeled immunocomplex. The beads are then re-suspended in a small volume (2 ml) of substrate, fluorescein di-β-D-galactopyranoside (FDG) and the mixture is partitioned into pL droplets such that most droplets have zero beads and a small percentage has one bead. The droplets are then loaded into a chamber in a monolayer to form droplet arrays. Images in three channels are obtained to identify the droplets, the beads, and the fluorescent product and thus the “on droplets”. Yamei Cai |

|
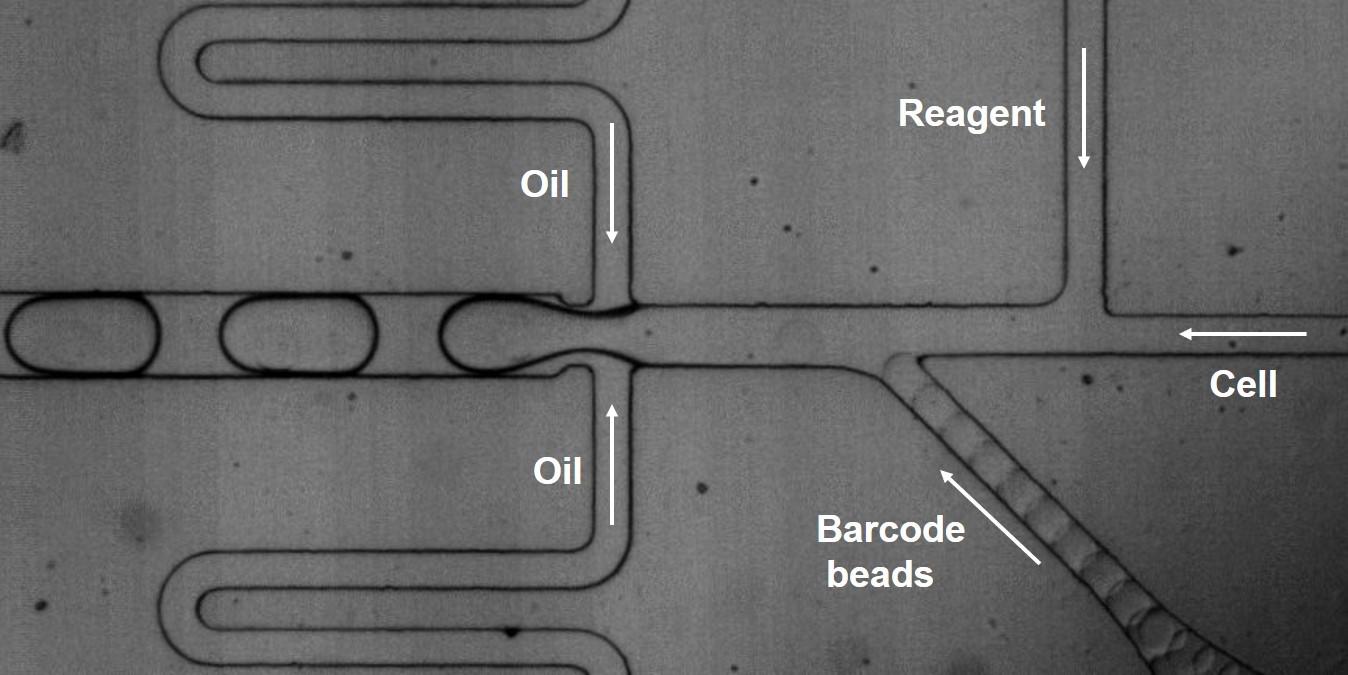
Integrating nanoplasmonic heating with ddPCR for rapid and high-sensitivity diagnostics: Digital droplet polymerase chain reaction (ddPCR), a method in which DNA is amplified and contained in a water-oil-based droplet, has become an attractive assay for clinical diagnostics as it provides high sensitivity to detect pathogenic genetic material. However, in order to expedite ddPCR for rapid diagnosis, we are integrating nanoplasmonic photothermal heating for ultrafast PCR thermocycling. To achieve this, we incorporate gold nanorods in the continuous solution in order to avoid their direct contact with PCR reagents inside the droplets, which would otherwise introduce inefficiencies in PCR amplification due to interactions of the gold nanorods with the proteins/enzymes. Overall, this system provides the basis for successfully integrating ultrafast PCR amplification via nanoplasmonic heating with highly-sensitive ddPCR detection. Jean Carlos Serrano Flores |
|
Droplet microfluidics for high-throughput single cell RNA sequencing: Single cell RNA sequencing (scRNA-seq) is becoming increasingly powerful for basic research, disease diagnosis and therapeutic development. Previously, our lab has co-developed the inDrop and Drop-seq platforms using droplet microfluidics for high-throughput scRNA-seq. Based on those platforms, we further improved the RNA detection sensitivity by developing a new chemistry to sequence the total RNA from single cells. Moreover, our new method is able to profile the transcriptome in single bacteria, which is previously inaccessible due to its lack of polyA tail on RNA transcripts, low RNA abundance and tough cell wells. This method will have broad impact on single-cell sequencing and microbiome research. Yongcheng Wang |
| Fast Droplet Digital Polymerase Chain Reaction (Fast ddPCR): ddPCR is a technology that uses discrete droplets to accurately quantify target nucleic acids in a sample. For example, we can use ddPCR to detect the nucleic acids of Coronavirus (COVID-19). Herein, we develop a medical instrument mainly including droplet microfluidics, thermal cycler and laser detector. As the core component of the instrument, the microfluidic chip integrates droplet generation, nucleic acid amplification and fluorescence detection. However, in the traditional method requires three instruments to achieve all the functions. Moreover, the whole detection process can be completed within 20 minutes and the efficiency is ten times the traditional dPCR instrument. And we are trying to realize the instrument to detect multiple samples simultaneously. Linbo Liu and Cui Wu | |
|
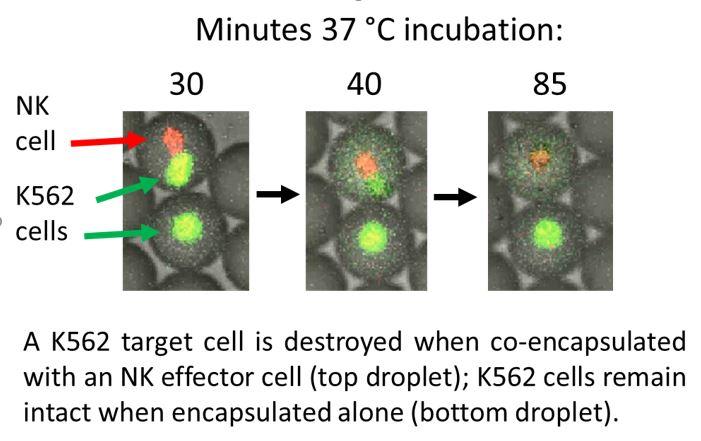
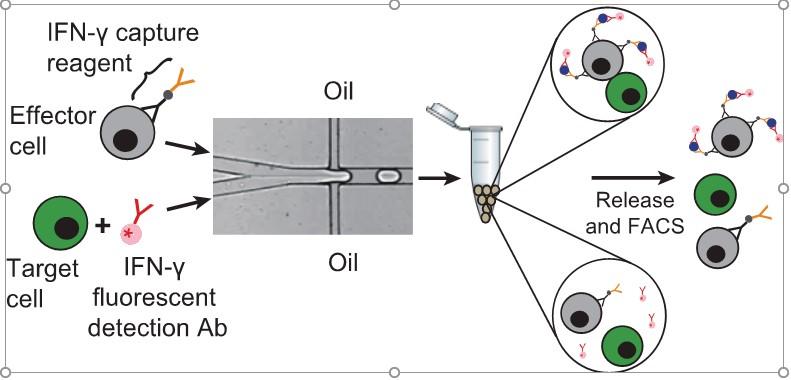
Droplet encapsulation improves accuracy of immune cell cytokine capture assays Quantification of cell-secreted molecules, e.g., cytokines, is fundamental to the characterization of immune responses. Cytokine capture assays that use engineered antibodies to anchor the secreted molecules to the secreting cells are widely used to characterize immune responses because they allow both sensitive identification and recovery of viable responding cells. However, if the cytokines diffuse away from the secreting cells, non-secreting cells will also be identified as responding cells. Here we encapsulate immune cells in microfluidic droplets and perform in-droplet cytokine capture assays to limit the diffusion of the secreted cytokines. We use microfluidic devices to rapidly encapsulate single natural killer NK-92 MI cells and their target K562 cells into microfluidic droplets. We perform in-droplet IFN-γ capture assays and demonstrate that NK-92 MI cells recognize target cells within droplets and become activated to secrete IFN-γ. Droplet encapsulation prevents diffusion of secreted products to neighboring cells and dramatically reduces both false positives and false negatives, relative to assays performed without droplets. After cells are released from the droplets, secreted cytokine remains captured onto secreting immune cells, enabling FACS-isolation of populations highly enriched for activated effector immune cells. Droplet encapsulation can be used to reduce background and improve detection of any single-cell secretion assay. Yuan Yuan |
| Combining microfluidics technologies with directed enzyme evolution: 1. to design a droplet microfluidic directed evolution system for selection of the optimal polymerase under various experimental conditions and 2. to better understand the sequence-function relation of polymerases by using machine learning techniques and potentially propose gene sequences of novel polymerases with high catalytic activity tailored for multiple adverse conditions. Xingge Zhang | |

|
Droplet Barcoding Test for Detection rare DNA Molecules: We encapsulates single DNA templates into drops, amplifies them by PCR, and then labels them with “barcodes” prior to sequencing them via massively-parallel sequencing platforms, such as the Illumina MiSeq. The computational fusion of the PCR product sequences yields an error-free sequence of each input DNA template. The process uses microfluidic technologies to synthesize the drops, encapsulate DNA, introduce reagents into drops, and prepare the amplified targeted molecules for sequencing. Ye Tao |
|
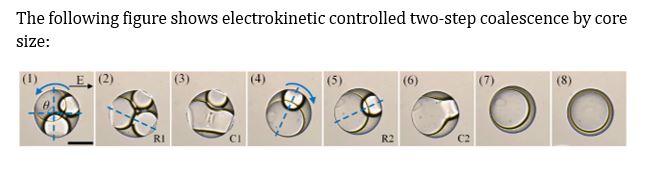
Electrokinetic Manipulation of Double-Emulsion Droplets: Double-emulsion dropelts are droplets of one or more liquids encased within a second immiscible liquid drops, which show great potential applications in microreactors, templates for 3D structures and functional materials and vessels for drugs etc. In many of these applications, cores in these double-emulsion drops have to be controlled coalescence or release accurately in the core-shell structure. We introduce electric field as a flexible tool to take up the task, because it has advantages of highly controllability, fast response and simple mechanism. Following realized the electrokinetic mechanism of the interaction between the electric field and double-emulsion droplet, we achieve the electrocoalescence of paired droplets and controlled rapid release of actives encapsulated in double-emulsion drops, and sequential electrocoalescence enabled two-step microreactions in triple-cored double-emulsion drops. More microfluidic devices with integrated functions are ongoing experiments. Yunkun Ren |
|
|
Microfluidics for single cell analysis of human antibody-secreting cells. Identification of new monoclonal antibodies able to target a specific epitope, for example on a virus capsid or on a cancer cell, is crucial for immuno-therapy development and vaccine design. The human body is an amazing source of antibodies: each human immune system contains >100 million different B cells, each producing many copies a unique antibody. Because each cell encodes only one type of antibody, an ability to test the activity of antibodies secreted by single cells will dramatically speed the identification of useful antibodies. To accomplish this, we have developed droplet microfluidic assay in which individual cells are encapsulated in 50pL micro-reactors containing all reagents to test the function of the secreted antibody. This assay allow us to isolate relevant B cells and to sequence the mRNA encoding the antibody. We are now increasing the accuracy and throughput of this assay by combining the single cell resolution of droplet microfluidics with the advanced optics and high-speed sorting of Fluorescence Activated Cell Sorting (FACS), to facilitate interrogation of a large repertoire of individual, non-immortalized cells. Using this technology we seek to identify monoclonal antibodies that could improve immuno-therapy in the fields of AIDS and cancer. Julie Brouchon |
|
|
Drop-Seq: High-throughput Single-cell RNA-Seq: Drop-based microfluidics have recently become a novel tool by providing a stable linkage between phenotype and genotype for high throughput screening. However, use of drop-based microfluidics for screening high-affinity peptide binders has not been demonstrated due to the lack of a sensitive functional assay that can detect single DNA molecules in drops. To address this sensitivity issue, we introduced in vitro two-hybrid system (IVT2H) into microfluidic drops and developed a streamlined mix-and-read drop-IVT2H method to screen a random DNA library. Drop-IVT2H was based on the correlation between the binding affinity of two interacting protein domains and transcriptional activation of a fluorescent reporter. A DNA library encoding potential peptide binders was encapsulated with IVT2H such that single DNA molecules were distributed in individual drops. We validated dropIVT2H by screening a three-random-residue library derived from a high-affinity MDM2 inhibitor PMI. The current drop-IVT2H platform is ideally suited for affinity screening of small-to-medium-sized libraries (10^3–10^6). It can obtain hits within a single day while consuming minimal amounts of reagents. Drop-IVT2H simplifies and accelerates the drop-based microfluidics workflow for screening random DNA libraries, and represents a novel alternative method for protein engineering and in vitro directed protein evolution. Naiwen Cui |
|
|
Microfluidic droplets for rapid isolation of individual cells with desired activity: We develop droplet-microfluidics assays to identify and isolate individual cells of interest. We have used a droplet-based fluorescence-concentration assay to isolate, from an input comprised primarily of cells that secrete irrelevant antibody, a fraction of living cells comprised almost entirely of cells secreting antibody against antigen of interest. We are currently developing assays to identify and select individual, living T-cells that recognize and kill specific target cells. Our ultimate goal is to commercialize these microfluidic methods. John Heyman. |
|
|
Droplet-based digital PCR: Digital PCR uses partitioning to provide a digital readout of the number of DNA strands present in a sample. Here, digital PCR is performed in droplets to increase sensitivity and reduce reagent use. In particular, the droplet digital PCR technique will be applied to studying low abundance gene mutations. Huidan Zhang |
|
|
Low-Concentration Quantification: Detection and quantification of low-concentration substances or microorganisms is difficult using conventional methods. We leverage the sequestration capabilities of droplets to create localized high concentrations in order to create an accurate measurement method. At present, we are able to perform accurate measurements in the attomolar range, and are working at increasing the sensitivity to improve this considerably. Jonathan Didier |
|
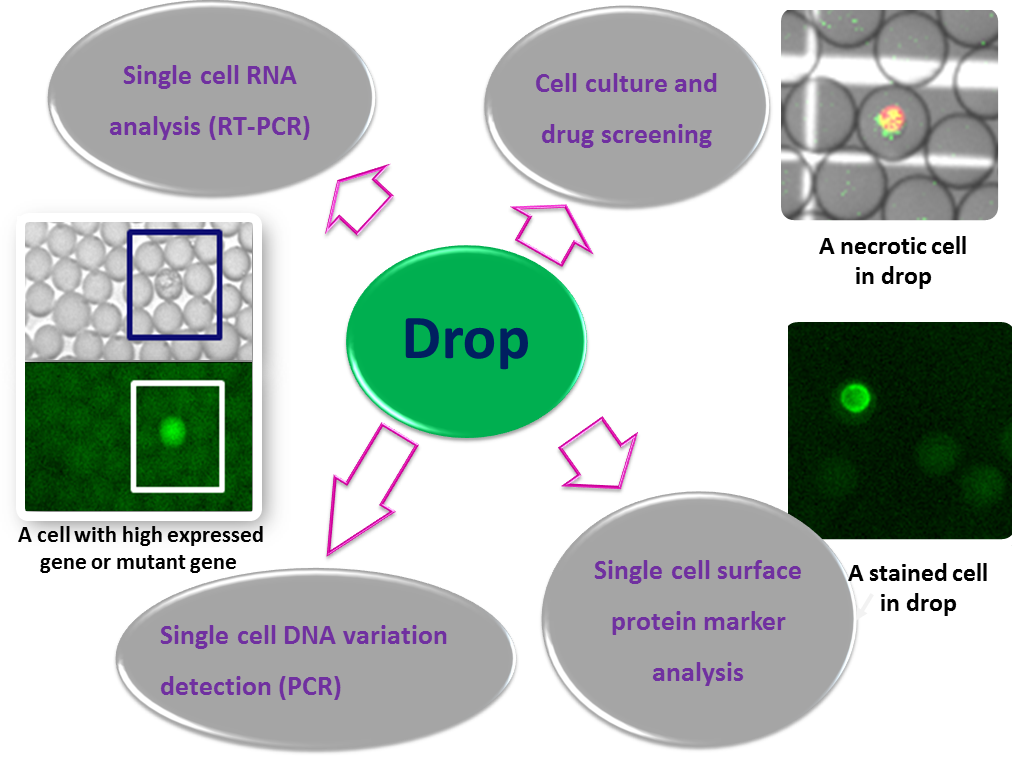
Single-Cell Polymerase Chain Reaction (PCR): High-throughput single cell-based screening can benefit considerably from drop-based microfluidics. This technique addresses the need for lower cost, shorter time, and higher sensitivity by using water-in-oil emulsion droplets to compartmentalize reactants into picoliter volumes. In our current research work, we test the biological reactions in drops at all the levels of single cells, including single-cell PCR to detect genomic variation, single-cell RT-PCR to analyze gene expression, single-cell surface protein marker analysis, and long-term cell culture. This promising platform provides a powerful tool to study single cells, thus allows us to explore the heterogeneity in development and tumorigenesis. Huidan Zhang |
|
|
Drops to Help Study the Brain: How do emotions, thoughts, or behaviors relate to physically observable properties of neurons? Fully answering this question may require first solving a technical challenge in studying the brain. How can we simultaneously measure the activity of billions (or more) individual neurons separated by distances from microns to centimeters? We apply micro- and milli-fluidic emulsions to this problem. In one project, we seek to engineer a DNA polymerase that could be used to record the voltage of many individual neurons simultaneously, a task that may be beyond the scope of optical, electronic, and magnetic recording methods. The recording medium would be DNA, and sequencing the DNA would read back the time series of neuronal pulses. We screen 100s-1000s of drops per second and confirm that we can detect fluorescent drops containing a single DNA polymerase gene, a step towards screening for a polymerase mutant that could record neuron activity into DNA. In another project, we encapsulate 0.1-1 mm clusters of human stem cells in drops that can gel and contain growth and differentiation factors. The stem cells differentiate into neurons, resulting in organoids, clusters of cells that resemble neocortical regions of the brain. It would be interesting to test in such organoids how a Ca2+ sensitive DNA polymerase records neuron activity by comparing recorded nucleotide sequences with microscopy recordings of Ca2+ dye fluorescence from the same organoid. Jesse Collins |
|
Cell Assays in Alginate Hydrogel Microparticles: I develop the high throughput method to encapsulate different cellular systems into alginate hydrogel microparticles. The alginate hydrogel platform has high biocompatibility allowing continuous cell culturing and high adaptability with high speed sorting using FACS. I further apply this technology platform to address 3 biomedical challenges: engineering a model of transmitted M. tuberculosis for vaccine testing, T cell receptor identification for cancer immunotherapy and cortical neuron differentiation for drug screening. Xu Zhang |
|
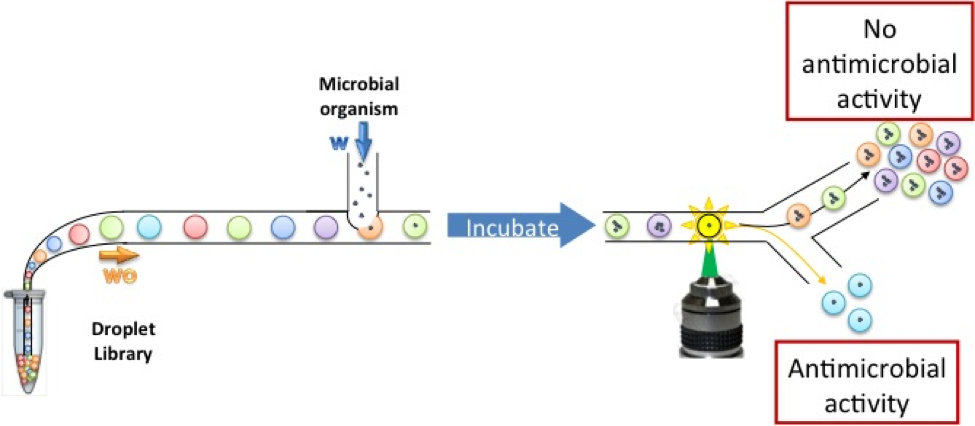
High throughput screening of active compounds using droplet microfluidics: We use droplet microfluidics to screen large libraries of compounds. Few picoliters of each compound is encapsulated in aqueous droplets and interacted with a target molecule or organism. By reducing the reagent volumes and reaction time, we can perform extremely high throughput screens at lower costs. ilke akartuna |

|
Elucidating Lipid Domains Function by Combinatorial Screening of Protein-Lipids Interactions: Membrane proteins are believed to require certain lipid compositions in order to incorporate in the lipid membrane, yet to date, we have limited tools to determine with which lipid domains a protein associates with. By the use of a large liposome library and microfluidics we can now determine the lipid mixtures a protein associates with. Most of the membrane proteins we studied so far, have shown to be highly specific to the lipid content in the liposomes. Roy Ziblat |
|
|
Next Generation Cell Purification Platform Using Acoustic Switching: Separating individual cells of interest from a mixed population of cells is of great importance and used in numerous applications in our daily life. For example, in planned parenthood; enabling sex selection of the child. Despite of the pure selective aspect; cell purification is a powerful tool in combination with i.e. adoptive cell therapy; optimizing the therapeutic outcome by selecting only the most vital and effective immune cells for cancer treatment. However, current standard methods such as fluorescence activated cell sorting (FACS) have their limitations in terms of biosafety, cost efficiency and the handling of specific types of samples. The focus of my work lies on the development of a microfluidics platform, which can overcome these limitations. I am particularly interested in using acoustics as a switching technology offering a universally usable, non-invasive and cost effective method to sort particles of interest, such as cancer cells, in a fluid stream of a microfluidic chip. Pascal Spink |
|
|
Novel Biomedical Applications Using Surface Acoustic Wave Microfluidics. Just as smart phone apps speak to our interests and desires for customization; providing a range of tools aggregated to a singular device, microfluidics, has become the mobile app analog for the biotechnology field. Despite providing applications such as cell sorting, reagent mixing, diagnostics, and more; microfluidics still remains limited. To circumvent these limitations microfluidics will need to be combined with other technologies. One promising union is that of surface acoustic wave technology and microfluidics. Dubbed acoustic microfluidics, this growing technology spans a number of disciplines in an attempt to address the shortcomings of microfluidics. My focus is to investigate different applications that may be possible for acoustic microfluidics such as developing microfluidic sensors, high-speed cell sorting, and single cell analysis applications. Kirk Mutafopulos |
|
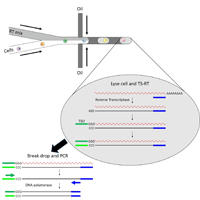
High-throughput and high-sensitivity single-cell RNA sequencing. Single-cell RNA sequence (scRNA-seq) is becoming a powerful technology to profile cell types in single-cell level, because the profile of the heterogeneous cells would provide invaluable information for diagnosis and treatments. Current scRNA-seq suffers only 10% recovery of total transcripts from a single cell and the inefficiency to recover low-abundance transcripts, such as T cell receptor (TCR) from T cell. My research goal is to develop a high-throughput and high-sensitivity scRNA-seq to increase the recovery of total transcripts and identify low-abundance transcripts of interest. To achieve my goal, droplet microfluidics together with template-switching reverse transcription polymerase chain reaction (TS-RT-PCR) are developed to recover the TCR transcripts from a single T cell. This technology would provide higher throughput and sensitivity to profile low-abundance transcripts without lose the sequencing power to sequence whole transcriptome. Kuo-Chan Hung |
|
|
Merging microfluidicis and metagenomics for novel high throughput virus discovery: This project is focused on developing the microfluidic platform for the detection, isolation, and complete sequencing of multiple types of viral genomes for use in standard molecular virology laboratories. For the experiments, total virion populations are isolated from the environmental sample, and deep sequencing and subsequent computational analysis detect the viral sequences present. We are aiming to isolate novel viruses of possible significance to human health and discover their whole genome sequences. Hee-Sun Han |

|
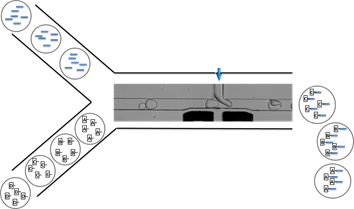
Evolution of small viral populations: A human viral disease can emerge when a single bird flu virus is able to infect a human host; characterizing the propagation of viral lineages is difficult but crucial for understanding, predicting and preventing the outburst of viral epidemics. A standard approach to probe this genetic propagation is by mutation accumulation experiments, in which a single lineage is propagated by plating a population of viruses, randomly picking a single colony to propagate, and repeating this process over time. Unfortunately, it is difficult to extract from these experiments enough information for a reliable characterization of an evolving species due to the small number of replicates collected and to the insufficient duration of the experiments. Using microfluidics we can perform millions of parallel mutation accumulation experiments in drops. To accomplish this, I developed an “evolution chip”, which passes millions of populations per hour from one drop where they replicated in the previous generation to a new drop containing fresh nutrients and hosts, where they will replicate in the next generation. When we co-encapsulated viruses and host cells in the same drop, viruses readily replicate in drops and smaller populations yield a broader clonal distribution. After propagating lineages of small populations in a new environment for 4 generations we find a much broader distribution of adapted clones compared to conventional passaging of large populations. Assaf Rotem |
|
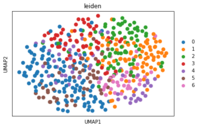
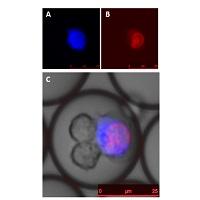
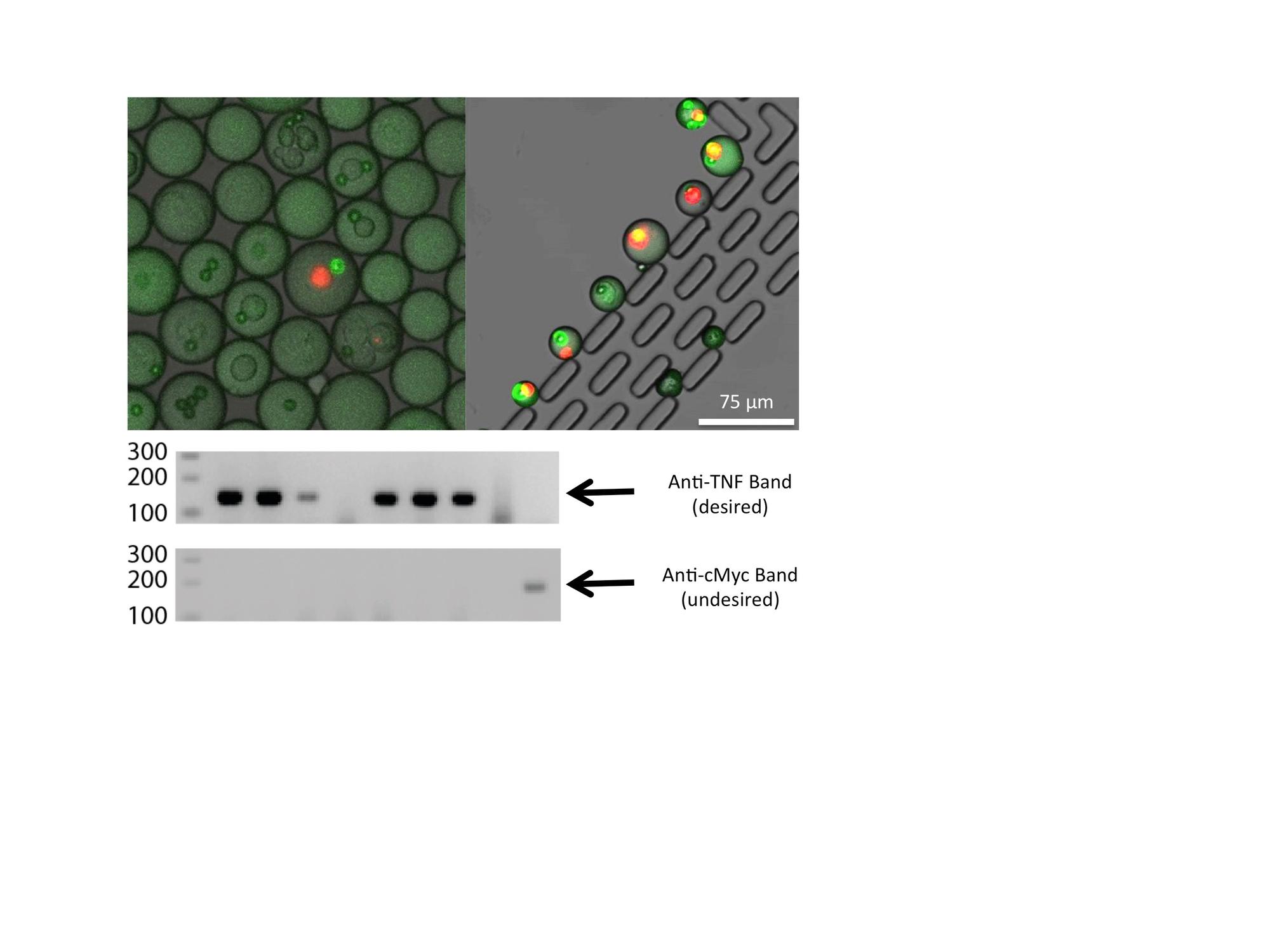
High throughput Single Cell Labeling (Hi-SCL): Cells of identical genetic origin develop to perform a variety of different functions in our body. The proteins encoded in the genome are not all expressed at the same level in each cell, leading to this cell-to-cell variation. Characterizing the cellular modifications that lead to this variation is key to understanding cellular differentiation in our body. However, the measurements for such modifications are currently performed over a population of cells and it is impossible to use them to study variations between cells in the same population. To perform these assays at the resolution of a single-cell we developed a new technique, called High-throughput Single Cell Labeling (Hi-SCL), where each cell is uniquely labeled in a drop. After labeling, drops are pooled and the emulsion is broken to make a single sample, which undergoes an epigenomic assay and deep-sequencing. Upon sequencing, the labels attached to the cellular fragments are used to associate each fragment with the cell of origin, enabling deconvolution of the epigenomic data to 100 profiles of single cells. These profiles are sufficient to identify two cell types that were pre-mixed in the sample, as described in Figure 3. Thus, Hi-SCL allows a sample of many cells to be assayed simultaneously, yet analyzed at single-cell resolution. Assaf Rotem |
|
Isolation of antigen-specific B-cells from primary cells. B-cells are immune cells that secrete antibody to fight pathogens in animals. The antibodies secreted by a single clone of B-cells, called monoclonal antibodies, are highly useful for both research and clinical applications. However, isolating B-cell clones that secrete antibodies that binds to a specific antigen using the traditional method of clonal expansion is inefficient and laborious. We develop a drop microfluidic platform to isolate B-cells based on the antibody they secrete. Using this platform we can isolate tens of different B-cells that secrete specific antibodies against a given antigen within one day. The antibody sequence can be subsequently retrieved by single-cell RT-PCR. This work will greatly improve the efficiency of antibody development and give better insights in antigen-antibody interactions. Ruihua Ding |
|
Low-abundance mutant KRAS detection. Constitutively active KRAS mutations have been found to be involved in various processes of cancer development. The poor detectable level of mutant type KRAS maintaining around 20% results from several problems like high background of view field under fluorescence light, and taqman hydrolyzes. Mutation detection methods with higher sensitivity will increase the possibility of choosing the correct individual therapy, and trying to realize low-abundance mutant KRAS detection is our goal. We test a new fluorescent probe for gene mutation detection using drop-based digital PCR and design a series of mixing experiment between KRAS mutations and wild type, and the minimum detectable ratio comes to 0.01%. Lexiang Zhang |
|
|
Drug susceptibility detection using double ddPCR. The goal of this project is to develop fast, sensitive and quantitative diagnostic methods for the identification of low-abundance bacterial pathogen in a bacterial infection where currently no diagnosis can be made. For the detection of low abundance mRNA to indicate drug susceptibility, we use a two rounds of drop-based digital PCR to perform a reliable detection. After the first round of drop-based digital PCR, pooled amplicons are re-encapsulated using a Poisson distribution to ensure <30% of droplets contain templates, are subjected to digital PCR and the bright droplets are counted by fluorescence detection. This method can expand the starting material without changing the original expression profile. Lexiang Zhang |
|

|
Effects of extract of Coreopsis tinctoria Nutt.on leukemia cells K562: Chinese herbal medicine attracts global attraction for the effective and safe effects. The Coreopsis tinctoria Nutt is a natural flower with many beneficial healthy effects. It is widely used as the flower tea and ethnic medicine in China. Recent studies have shown that the extract from Coreopsis tinctoria flower possess various biological activities. Our study provides the evidence that the extract from the Coreopsis tinctoria can be the potential candidate to treat leukemia.The mechanism might be inducing tumor cell apoptosis.The merit of the combination of Coreopsis tinctoria combined with Imatinib in inhibition luekemia carcinoma in vitro based on microfluidic chip technology will be further studied next step.Xinmei Chen |
|
Circulating Tumor Cells Detection: Circulating tumor cells are kinds of cancer cells that shed from the primary tumor into the blood stream. By CTCs enumeration and subtype classification, we can monitor ongoing progress of the tumor in a non-invasive manner. Herein, we develop a CTCs detection and subtype classification method based on the droplet microfluidics. First encapsulate the blood specimen into millions of droplets. Second, lysis the cells in the droplets, using multiplex fluorescent PCR to detect the specific mutations. At last, record the fluorescent intensity of each droplets, and calculate the CTCs number. Jidong Wang and Wenwen Chen |
|
|
