Single-cell analysis and screening using microfluidics
Droplet microfluidic systems have proven to be valuable tools for single-cell analysis, complex enzymatic assays, disease diagnostics, drug screening or directed evolution experiments. Due to reduced reaction volumes, unique liquid handling capabilities and increased analytical sensitivity microfluidic systems bring a new standard in high-throughput screens. A particular advantage is that droplets provide a unique way of linking genotype with phenotype through compartmentalization rather than a chemical bond. Moreover, the secreted molecules remain entrapped inside the droplet together with a cell that produce them. Since the reaction volumes are in the range of tens of picoliters the time necessary to achieve detectable protein concentration occurs within few minutes instead of hours that are more typical to microtiter plates screens. The microfluidic fluorescence activated droplets sorters can then be applied to identify and sort entire reaction vessels containing individual cells. Compartmentalized cells can be lysed and their genetic information recovered after DNA or RNA amplification inside the droplets. Using microfluidic tools we are working on various biological applications:
- Screening of single-cells producing therapeutic antibodies
- Integrated microfluidic platform for long-term cell cultivation, monitoring and analysis
- Single-cell transcriptomics
Screening of single-cells producing therapeutic antibodies
This work is performed together with John Heyman. Assays such as ELISA, routinely used to screen for antibody binding in microtitre-plates, are relatively slow (requires 5-7 days) and have limited throughput. Therefore, it is very appealing to develop droplet microfluidic systems to select antibody-secreting cells. For this purpose we developed novel binding assay that requires no washing step and is based on the concentration of fluorescence signal on a bead co-encapsulated with each cell (Figure 1). In this assay, the secreted antibodies are captured on the bead surface and detected using a fluorescently-labeled secondary antibody. As a result of this interaction, the fluorophore in the droplet becomes concentrated on the bead generating a clearly distinguishable signal. Finally, droplets containing fluorescent-beads and co-encapsulated cells are sorted using a microfluidic sorter.
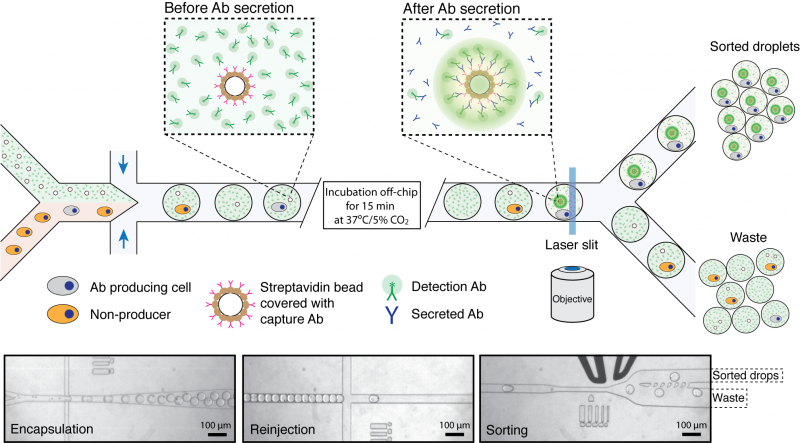
Figure 1. Schematics of microfluidic system for screening of antibody producing cells. Cells are introduced into a microfluidic device together with a bead suspension and fluorescently-labeled goat detection antibodies (green) and beads coated with goat anti-mouse-Fc capture antibodies (pink). After encapsulation and incubation for 15 min at 37ºC/5%CO2, those beads that are co-encapsulated with an antibody producing cell become highly fluorescent due to the capture of secreted antibodies and binding of the fluorescent label. The emulsion is then introduced into a second microfluidic device and droplets containing fluorescent beads are sorted using a fluorescence activated droplet sorter. The three micrographs show i) co-encapsulation of cells with beads, ii) droplet reinjection after incubation off-chip and iii) droplet sorting. See Nature Protocols 2013, Vol.8, No.5, p870 for more details.
Integrated microfluidic platform for long-term cell cultivation, monitoring and analysis
This work is performed together with Prof. Andrew Murray group. We are developing the microfluidic device that allows production thousands of picoliter volume bioreactors, trap them into specially designed jackets and keep them in place over extended periods of time. Due to large number of bioreactors (~105) on a chip we can entrap large population of cells and then monitor individual cell growth and response over selected period of time using automated microscope. We inject the sample containing genetically identical yeasts together with necessary ingredients (e.g. sucrose, glucose) and compartmentalize cells into 50 pL bioreactors. We can adjust the size of bioreactors from 50 to 200 pL which gives us flexibility in experimental set-up. We inject different populations of yeasts and monitor their mutual interactions and adaptation in each bioreactor separately. The compartmentalization of cells is achieved at the flow-focusing junction, which brakes fluid into micro-droplets. These micro-reactors are then moving downstream the flow and get trapped inside specially designed channels (Figure 2). Once trapped, bioreactors can be kept at dedicated position for days and multiple cells can be followed with automated microscope. Using this type of microfluidic device we can analyze large number of cells under conditions that are hard to achieve in bulk. For example, we can use limited amount of nutrition and/or carbon source to determine how cells respond and adapt over multiple generations. We are currently investigating cell adaptation to limited amount of glucose and sucrose.
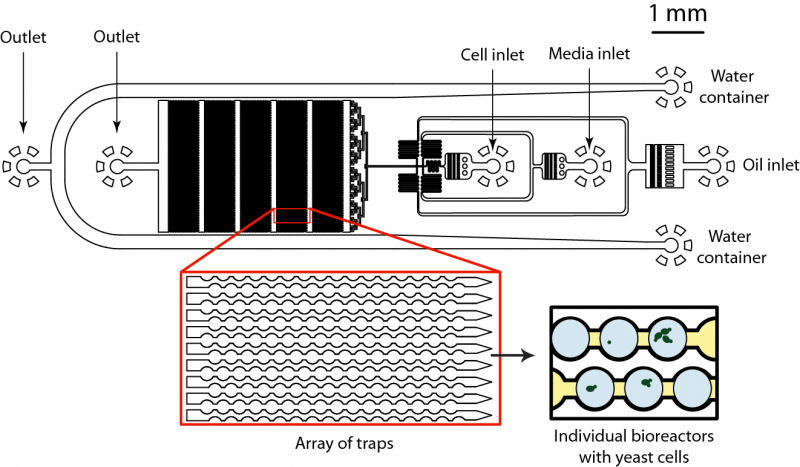
Fig 2. Microfluidics device for analysis and monitoring of large number of bioreactors on a chip. The microfluidic device consists of two sample inlets and one inlet for continuous phase. Each bioreactor of 50 pL in volume is created by dispersing injected liquid at the flow-focusing junction. Then each bioreactor flows downstream the flow and is trapped in specially designed pockets that keep them at place and allows long-term monitoring and analysis. The cellular growth and gene expression is monitored with automated microscope.
Single cell transcriptomics
This work is performed in collaboration with Allon Klein at Harvard Medical School and with Ilke Alkartuna from our group. Single-cell sequencing has been announced by the Editorial of Nature Methods as a method of the year 2014. Indeed, recent breakthrough in single-cell sequencing promises to bring new level of understanding about the cell heterogeneity in biology. Many techniques, however, although powerful and efficient can analyze only limited amount of cells, with current limit being ~ 100-1000. We are developing a novel approach for decoding the whole transcriptome of large number (>10.000) of single-cells. For that purpose we are encapsulating individual cells into microfluidic droplets together with DNA barcodes that, after cell lysis and reverse transcription steps, get incorporated into a newly synthesized cDNA. By sequencing pooled cDNA product we are aiming at decoding the gene expression levels of each cell.
Linas Mazutis is supported by Marie Curie International Outgoing Fellowship (300121).
